By sequencing an enormous amount of data, a group of hundreds of researchers has gained new insights into how flowers evolved on Earth.
Almost every plant we eat has a flower, and flowering plants populate every corner of the planet. But many questions remain about how and when this vast group emerged throughout the history of life on Earth.
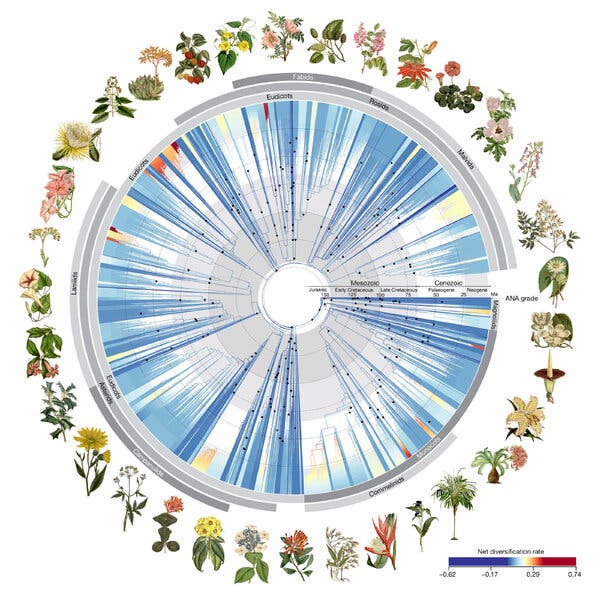
Now, after a heroic DNA sequencing effort, a collaboration involving hundreds of scientists has created a new family tree for flowering plants. Comparing gene sequences from more than 9,500 species — many of them dried specimens preserved in museums — scientists have sketched important branching points in the evolution of flowering plant life. In a study published in April in the journal Nature, the data they present suggests that more than 80 percent of major modern flowering plant lineages originated in a sudden burst of invention that began around 150 million years ago, in the late Jurassic Period.
Previous evolutionary trees of plants built by scientists often used the genome of the chloroplast, the organelle that allows plants to perform photosynthesis. These genomes could be sequenced with older methods. But scientists could not be confident that the patterns they showed were the same as what might be revealed by the plant’s primary genome, stored in the cell’s nucleus and more difficult to study.
Then, five years ago, another scientific collaboration published detailed information about more than 1,100 plant species’ nuclear genomes. That allowed the team behind the Nature paper to design new tools for sequencing nuclear genes from a huge variety of flowering plants, said William Baker, who leads the Kew Gardens Tree of Life Initiative and is an author of the new paper.
They used the tools on living plants, but the team also reached out to institutions in 48 countries with collections of dried plants to get samples of rare specimens. Four of the plants included in the analysis are already extinct, including the Guadalupe Island olive, which was sequenced using a dried sprig from 1875. In the end, the team used data from about 60 percent of all modern genera of plants.
As they put the new evolutionary tree together, they found that it confirmed many of the relationships suggested by trees built from chloroplasts. However, there were surprises: The new data reshuffled the relationships of a number of plant groups, and some individual species were reclassified.
figcaption[data-testid=”photoviewer-children-caption”] {
margin: 12px auto 0 auto;
padding-left: 13px;
padding-right: 13px;
text-align: center;
}
figcaption class=”css-1ifeaca e3rygrp0″ {
margin: 12px auto 0 auto;
padding-right: 13px;
padding-left: 13px;
text-align: center;
}
/* TOP AD CODE BELOW */
#top-wrapper, sponsor-wrapper { display: none; }